Revealed: the protein 'spike' that lets the 2019-nCoV coronavirus pierce and invade human cells

Researchers in the United States have unveiled the structure of the “spike protein” of 2019-nCoV – the virus behind the current coronavirus disease outbreak.
Despite the fact that researchers have already pieced together the virus’s genetic sequence, the World Health Organisation has warned that a vaccine is still 18 months away.
Read more: Here's why the WHO says a coronavirus vaccine is 18 months away
But knowing the structure of the virus’s spike protein gives us crucial information about exactly how the virus infects host cells. This could be a vital piece of the puzzle in making the hoped-for vaccine a reality.
What is a spike protein?
A viral spike protein is like a key that “unlocks the door” to gain access to the cells of a specific host – humans, in this case. To understand how to deal with 2019-nCoV, we first need to understand what this key looks like, and what “keyhole” it targets on human cells. This is exactly what the new paper, published overnight in Science, is all about.
The researchers, led by Jason McLellan of the University of Texas at Austin, defined the structure of 2019-nCoV’s spike protein using a technique called cryogenic electron microscopy, or “Cryo-EM”. This involves cooling the protein to below -150℃, so that it crystallises and then its structure can be determined with near-atomic resolution.
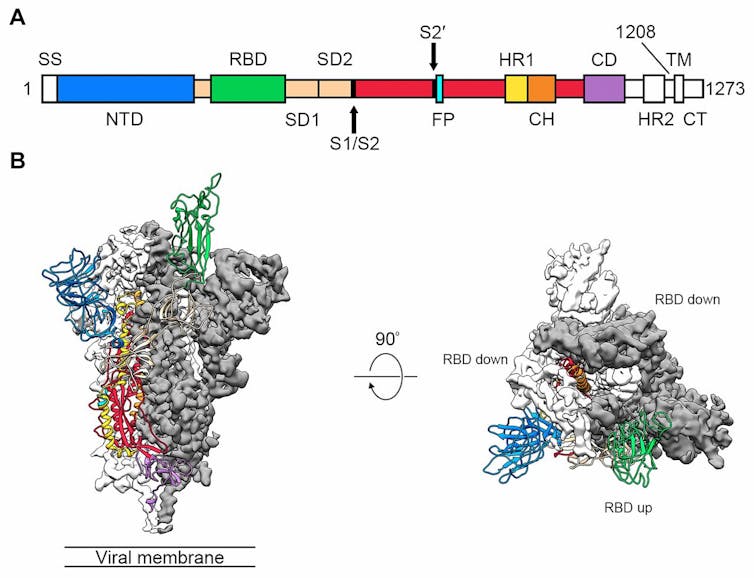
They also identified the “keyhole”, the host cell receptor: it is a human protein called angiotensin converting enzyme 2 (ACE2). This is the same human receptor protein targeted by the earlier SARS coronavirus.
But, disturbingly, the researchers found that 2019-nCoV binds to ACE2 with much higher affinity (10-20 times higher!) than SARS. In other words, 2019-nCoV’s “key” is a lot “stickier” than the SARS one. It’s like a SARS “key” covered in superglue. This means that once it’s in the lock, it’s far less likely to be shaken loose and is therefore presumably more effective at invading our cells.
So what about a vaccine?
The researchers reasoned that, given that both viruses attack the same protein on human cells, it would be worth seeing whether the already available antibodies against SARS-CoV would work against 2019-nCoV. Unfortunately, they didn’t work.
This means we still have to wait for a stronger solution to this problem. Perhaps this is a reflection of the ongoing “arms race” between humans and viruses. We have stronger weapons now, thanks to scientific advances, but our enemies are gaining strength too – now they are using superglue against us!
Read more: We're in danger of drowning in a coronavirus 'infodemic'. Here's how we can cut through the noise
Globally, the competition is heating up to hunt for the best anti-2019-nCoV vaccine. But as the old Chinese proverb says, “distant water can’t put out a nearby fire”. The earliest clinical trials to test a suitable vaccine will not be available until several months or even a year after a candidate vaccine is identified, and the global coronavirus outbreak may well be controlled by then.
The discovery of the 2019-nCoV spike protein structure therefore represents both good news and bad. The good news is now we know what it looks like, it will be easier to find the most suitable weapon against the virus. The bad news is the enemy is much stronger than we thought, and our current ammunition depot doesn’t have anything efficient against it.![]()
Jianling Xie, Postdoctoral Scientist, South Australian Health & Medical Research Institute
This article is republished from The Conversation under a Creative Commons license. Read the original article.